I-SceI
Type
Wild-Type
Target Sequence
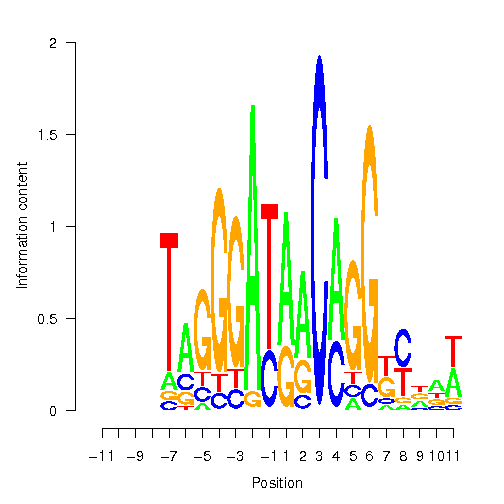
| -11 | -10 | -9 | -8 | -7 | -6 | -5 | -4 | -3 | -2 | -1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| A | C | G | C | T | A | G | G | G | A | T | A | A | C | A | G | G | G | T | A | A | T |
FASTA Sequence
MKNIKKNQVMNLGPNSKLLKEYKSQLIELNIEQFEAGIGLILGDAYIRSRDEGKTYCMQFEWKNKAYMDHVCLLY
DQWVLSPPHKRERVNHLGNLVITWTAQTFKHQAFNKLANLFIVNNKKTIPNNLVENYLTPMSLAYWFMDDGGKWD
YNKNSTNKSIVLNTQSFTFEEVEYLVKGLRNKFQLNCYVKINKNKPIIYIDSMSYLIFYNLIKPYLIPQMMYKLP
NTISSETFLK
DQWVLSPPHKRERVNHLGNLVITWTAQTFKHQAFNKLANLFIVNNKKTIPNNLVENYLTPMSLAYWFMDDGGKWD
YNKNSTNKSIVLNTQSFTFEEVEYLVKGLRNKFQLNCYVKINKNKPIIYIDSMSYLIFYNLIKPYLIPQMMYKLP
NTISSETFLK
Crystal Structure
| PDB File |
| PyMOL File |
Position Weight Matrix
|
|
|
Specificity Changing Mutations
| Amino Acid Mutations (?) | Specificity Changes | Reference |
| K86R, G100T | +4A to +4C, +4C to +4C | 1 |
| K86R, G100C | +4A to +4C, +4C to +4C | 1 |
| K86R, G100S | +4A to +4C, +4C to +4C | 1 |
| W149G, D150C, N152K | -7T to -7C | 2 |
Additional Information
Author: Barry StoddardNotes:
Date Added: 2011-10-18