2011
Target Site Interpretation
For each homing endonuclease, a list of candidate target sequences are returned from any of the search procedures. Each target site is displayed with information including the gene in which it is found, the position and orientation within the gene, the target sequence itself, and a score for the match. These search results are ordered by their score, which is calculated by multiplying matching frequencies within the underlying PWM across the target site. These scores may be compared with the score of the natural target site which is displayed first for each homing endonuclease.
For each candidate target site, nucleotides across the site from the -11 to +11 positions are displayed. Bases that match the wild type site are in upper case and mismatches in lower case (often in red). Matching nucleotides from the central four bases (-2 to +2) are upper case and in blue. These central four nucleotides are often considered the most difficult to engineer away from in engineering experiments.
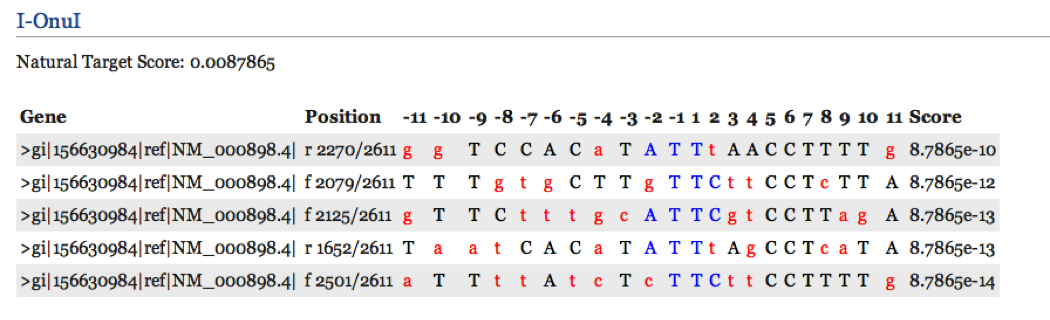
Individual bases within each target site link to information that may assist in the reengineering process. For example, clicking on the I-MsoI -6 position will return a list of mutations used to change specificity from -6C to +6G and +6A to +6 C. References describing these positions are displayed below the mutations. Additionally, information available for a module that could help engineering of the specific site will be displayed if it is available.