I-CreI
Type
Wild-Type
Target Sequence
| -11 |
-10 |
-9 |
-8 |
-7 |
-6 |
-5 |
-4 |
-3 |
-2 |
-1 |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
| G |
A |
A |
A |
A |
C |
G |
T |
C |
G |
T |
G |
A |
G |
A |
C |
A |
G |
T |
T |
T |
C |
FASTA Sequence
MNTKYNKEFLLYLAGFVDGDGSIIAQIKPNQSYKFKHQLSLTFQVTQKTQRRWFLDKLVDEIGVGYVRDRGSVSD
YILSEIKPLHNFLTQLQPFLKLKQKQANLVLKIIEQLPSAKESPDKFLEVCTWVDQIAALNDSKTRKTTSETVRA
VLDSLSEKKKSSP
1 6 11 16 21 26 31 36 41 46 51 56 61 66 71
MNTKYNKEFLLYLAGFVDGDGSIIAQIKPNQSYKFKHQLSLTFQVTQKTQRRWFLDKLVDEIGVGYVRDRGSVSD
76 81 86 91 96 101 106 111 116 121 126 131 136 141 146
YILSEIKPLHNFLTQLQPFLKLKQKQANLVLKIIEQLPSAKESPDKFLEVCTWVDQIAALNDSKTRKTTSETVRA
151 156 161
VLDSLSEKKKSSP
Crystal Structure
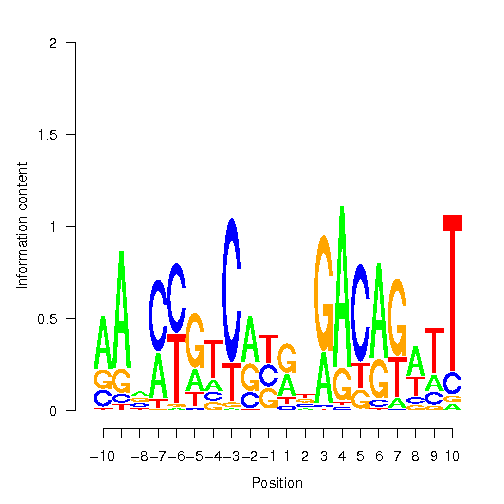
Position Weight Matrix
|
|
| pos | A | C | G | T |
| -11 |
0.25 |
0.25 |
0.25 |
0.25 |
| -10 |
1.00 |
0.31 |
0.35 |
0.04 |
| -9 |
1.00 |
0.07 |
0.20 |
0.05 |
| -8 |
1.00 |
0.71 |
0.87 |
0.39 |
| -7 |
0.63 |
1.00 |
0.01 |
0.11 |
| -6 |
0.03 |
1.00 |
0.03 |
0.98 |
| -5 |
0.40 |
0.04 |
1.00 |
0.25 |
| -4 |
0.28 |
0.22 |
0.21 |
1.00 |
| -3 |
0.01 |
1.00 |
0.03 |
0.26 |
| -2 |
1.00 |
0.34 |
0.59 |
0.01 |
| -1 |
0.01 |
0.77 |
0.71 |
1.00 |
| 1 |
0.75 |
0.15 |
1.00 |
0.22 |
| 2 |
0.43 |
0.84 |
0.89 |
1.00 |
| 3 |
0.44 |
0.02 |
1.00 |
0.01 |
| 4 |
1.00 |
0.02 |
0.20 |
0.02 |
| 5 |
0.00 |
1.00 |
0.19 |
0.26 |
| 6 |
1.00 |
0.07 |
0.41 |
0.02 |
| 7 |
0.12 |
0.01 |
1.00 |
0.51 |
| 8 |
1.00 |
0.21 |
0.17 |
0.63 |
| 9 |
0.35 |
0.27 |
0.09 |
1.00 |
| 10 |
0.04 |
0.14 |
0.04 |
1.00 |
| 11 |
0.25 |
0.25 |
0.25 |
0.25 |
|
Amino Acid Contact Modules
| Module | Amino Acid Contact Modules (?) |
| -11 → -9: | Lys 33, Gln 38 |
| -8 → -6: | Gln 26, Lys 28 |
Key
|
Experiment |
|
Crystal Structure |
|
Sequence Alignment |
| * |
Uncertain |
Specificity Changing Mutations
| Amino Acid Mutations (?) |
Specificity Changes |
Reference |
|
Y33H
|
-10A to -10G, +10T to +10C
|
1
|
|
Y33R
|
-10A to -10G, +10T to +10C
|
1
|
|
Y33C
|
-10A to -10T, +10T to +10A
|
1
|
|
Q26A
|
-6C to -6A, +6A to +6T
|
2
|
|
Q26C
|
-6C to -6G, +6A to +6C
|
2
|
|
Q26C, Y66R
|
-6C to -6G, +6A to +6C
|
2
|
| 1 |
Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. (2002) Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Research, 30, 3870-3879. |
| 2 |
Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Stoddard BL, Seligman LM. (2004) Isolation and characterization of new homing endonuclease specificities at individual target site positions. Journal of Molecular Biology, 342, 31-41. |
