I-LtrI
Type
Wild-Type
Target Sequence
| -11 | -10 | -9 | -8 | -7 | -6 | -5 | -4 | -3 | -2 | -1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| A | A | T | G | C | T | C | C | T | A | T | A | C | G | A | C | G | T | T | T | A | G |
FASTA Sequence
FPVQARNDNISPWTITGFADAESSFMLTVSKDSKRNTGWSVRPRFRIGLHNKDVTILKSIREYLGAGIITSDKDA
RIRFESLKELEVVINHFDKYPLITQKRADYLLFKKAFYLIKNKEHLTEEGLNQILTLKASLNLGLSEELKEAFPN
TIPAEKLLVTGQEIPDSNWVAGFTAGEGSFYIRIAKNSTLKTGYQVQSVFQITQDTRDIELMKNLISYLNCGNIR
IRKYKGSEGIHDTCVDLVVTNLNDIKEKIIPFFNKNHIIGVKLQDYRDWCKVVTLIDNKEHLTSEGLEKIQKIKE
GMNRGRSL
RIRFESLKELEVVINHFDKYPLITQKRADYLLFKKAFYLIKNKEHLTEEGLNQILTLKASLNLGLSEELKEAFPN
TIPAEKLLVTGQEIPDSNWVAGFTAGEGSFYIRIAKNSTLKTGYQVQSVFQITQDTRDIELMKNLISYLNCGNIR
IRKYKGSEGIHDTCVDLVVTNLNDIKEKIIPFFNKNHIIGVKLQDYRDWCKVVTLIDNKEHLTSEGLEKIQKIKE
GMNRGRSL
Crystal Structure
| PDB File |
| PyMOL File |
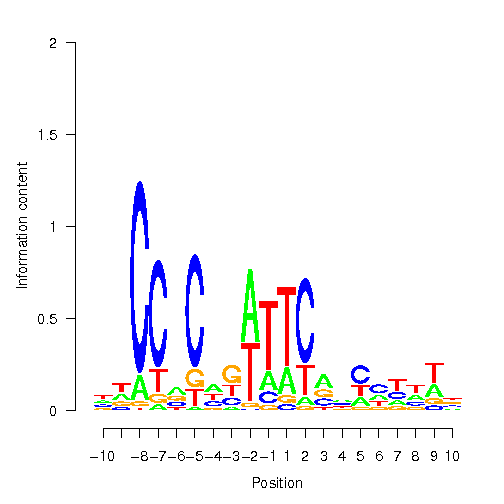
Position Weight Matrix
|
|
|
Amino Acid Contact Modules
| Module | Amino Acid Contact Modules (?) |
| -11 → -9: | Asp 39, Lys 41, Arg 42, Asn 43, Thr 44, Gly 45, Ser 47 |
| -8 → -6: | Met 33, Thr 35, Ser 37, Arg 49, Arg 51, Ile 75, Thr 77, Arg 85, Glu 87 |
| -6 → -4: | Ser 31, Met 33, Arg 53, Gly 55, Thr 77, Arg 83, Arg 85 |
| -5 → -3: | Ser 31, Arg 53, Gly 55, Ile 80*, Asp 81*, Arg 83, Arg 85 |
| +3 → +5: | Ser 186, Tyr 188, Gln 208, Thr 210, Arg 234, Cys 246, Asp 248 |
| +4 → +6: | Tyr 188, Arg 190, Val 206, Gln 208, Asn 230, Arg 232, Arg 234, Val 250 |
| +6 → +8: | Arg 190, Ala 192, Gln 202, Gln 204, Val 206, Asn 230, Thr 252 |
| +9 → +11: | Ala 186, Asn 194*, Thr 196*, Leu 197, Lys 198, Thr 199, Gly 200, Gln 202 |
Key
| Experiment | |
| Crystal Structure | |
| Sequence Alignment | |
| * | Uncertain |
Additional Information
Author: Barry StoddardNotes:
Date Added: 2012-01-30