I-AniI / I-OnuI
Type
Chimera
Target Sequence
| -11 | -10 | -9 | -8 | -7 | -6 | -5 | -4 | -3 | -2 | -1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| C | T | G | A | G | G | A | G | G | T | T | T | C | A | A | C | C | T | T | T | T | A |
FASTA Sequence
YMSRRESINPWILTGFADAEGYFSLTIKNNNKSSVGYYTTLEFGIELSNKDKSILENIQSTWKVGIISNRGINMV
ALRVRRKEDLKVIIDHFEKYPLITQKLGDYMLFKQAFCVMENKEHLKINGIKELVRIKAKLNWGLTDELKKAFPE
IISKERSLINKNIPNFKWLAGFTSGEGCFFVNLIKSKSKLGVQVQLVFSITQHIKDKNLMNSLITYLGCGYIKEK
NKSEFSWLDFVVTKFSDINDKIIPVFQENTLIGVKLEDFEDWCKVAKLIEEKKHLTESGLDEIKKIKLNMNKGRV
F
ALRVRRKEDLKVIIDHFEKYPLITQKLGDYMLFKQAFCVMENKEHLKINGIKELVRIKAKLNWGLTDELKKAFPE
IISKERSLINKNIPNFKWLAGFTSGEGCFFVNLIKSKSKLGVQVQLVFSITQHIKDKNLMNSLITYLGCGYIKEK
NKSEFSWLDFVVTKFSDINDKIIPVFQENTLIGVKLEDFEDWCKVAKLIEEKKHLTESGLDEIKKIKLNMNKGRV
F
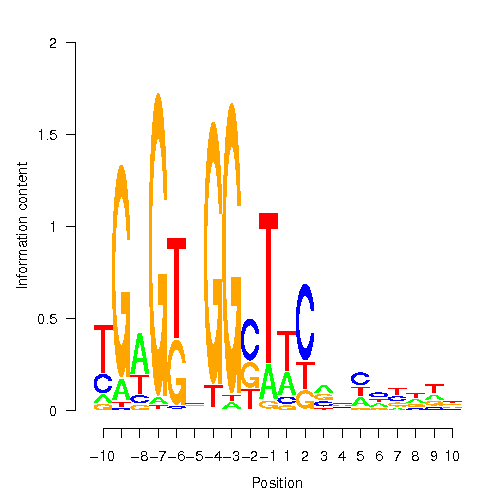
Position Weight Matrix
|
|
|
(-) Endonuclease Amino Acid Contact Modules
| Module | Amino Acid Contact Modules (?) |
| -11 → -9: | Tyr 27 |
| -8 → -6: | Ser 20, Thr 22, Gly 25*, Thr 29, Glu 31, Ile 55, Ser 57, Arg 70, Arg 72 |
| -6 → -4: | Tyr 18, Ser 20, Gly 33, Glu 35, Ser 57, Arg 59*, Ala 68, Arg 70 |
| -5 → -3: | Tyr 18, Gly 33, Glu 35, Arg 59*, Glu 65*, Met 66, Ala 68, Arg 70 |
(+) Endonuclease Amino Acid Contact Modules
| Module | Amino Acid Contact Modules (?) |
| +3 → +5: | Cys 180, Phe 182, Ser 201, Thr 203, Lys 227, Lys 229, Trp 234, Asp 236 |
| +4 → +6: | Phe 182, Asn 184, Val 199, Ser 201, Tyr 223, Lys 225, Lys 227, Val 238 |
| +6 → +8: | Asn 184, Ile 186, Gln 195, Gln 197, Val 199, Tyr 223, Thr 240 |
| +9 → +11: | Ile 186, Ser 188, Lys 189, Ser 190, Lys 191, Leu 192, Gly 193, Gln 195 |
Key
| Experiment | |
| Crystal Structure | |
| Sequence Alignment | |
| * | Uncertain |
(-) Endonuclease Specificity Changing Mutations
| Amino Acid Mutations (?) | Specificity Changes | Reference |
| K24N, T29K | -8A to -8G | 1 |
| Y18W, E35K, R61Q | -3G to -3C | 1 |
| K24N, G25R, T29I | -9G to -9C | 1 |
| T29S, E31R, R70L | -6G to -6C | 1 |
| 1 | Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL, Baker D. (2009) Exploitation of binding energy for catalysis and design. Nature, 461, 1300-1304. |